Jomol P. Mathew, PhD; Robert N. Golden, MD
WMJ. 2022;121(4):333-334
When COVID-19 was spreading throughout the world, health care providers and researchers needed rapid access to data that were emerging at lightning speed. Fortunately, we were able to leverage currently available data analytics and digital technologies in our battle against COVID-19. Electronic health records (EHR), which became more widespread following enactment of the American Recovery and Reinvestment Act of 2009,1 allowed iterative studies of symptoms and patient responses to treatments, while mobile technologies assisted in tracking the spread of the virus. Video-based technologies provided remote communication between providers and patients, and social media and the internet helped disseminate information. These developments likely contributed to the vastly reduced death toll (6.58 million worldwide as of November 11, 2022)2 compared to the 1918-1919 influenza pandemic (approximately 50 million worldwide).3 The COVID-19 pandemic radically changed our perspectives on how to conduct research and clinical care, and it set the stage for data-driven biomedical research and clinical practice.
Real-World Data in Biomedical Research and Health Care
The development of disease is influenced by a person’s genetics, exposome (lifetime environmental exposures), and interactions between them. Advances in human genome sequencing;4 high-throughput multi-omics;5 and computational, geospatial, and digital technologies have created unprecedented opportunities for studying genetic factors and the exposome in relation to risk for disease. While effective prevention, treatment, and management of diseases depend on these factors, relevant data from multiple perspectives are not often obtained, let alone integrated. The vastness, heterogeneity, and sparse linkage among data generated over a patient’s life course, in turn, limit the ability to integrate and analyze data in a comprehensive and timely way. It is imperative that we find ways to effectively utilize large biomedical data sets.
Following the lessons learned from the pandemic, and with the release of the US Food and Drug Administration’s new guidelines for using real-world data,6 universities and academic medical centers must develop the capacity to conduct multidisciplinary, data-driven research. The University of Wisconsin School of Medicine and Public Health (SMPH) has launched several initiatives to advance research and the translation of findings to clinical care. Here, we highlight a few examples.
The Wisconsin Real-World Data Collaborative
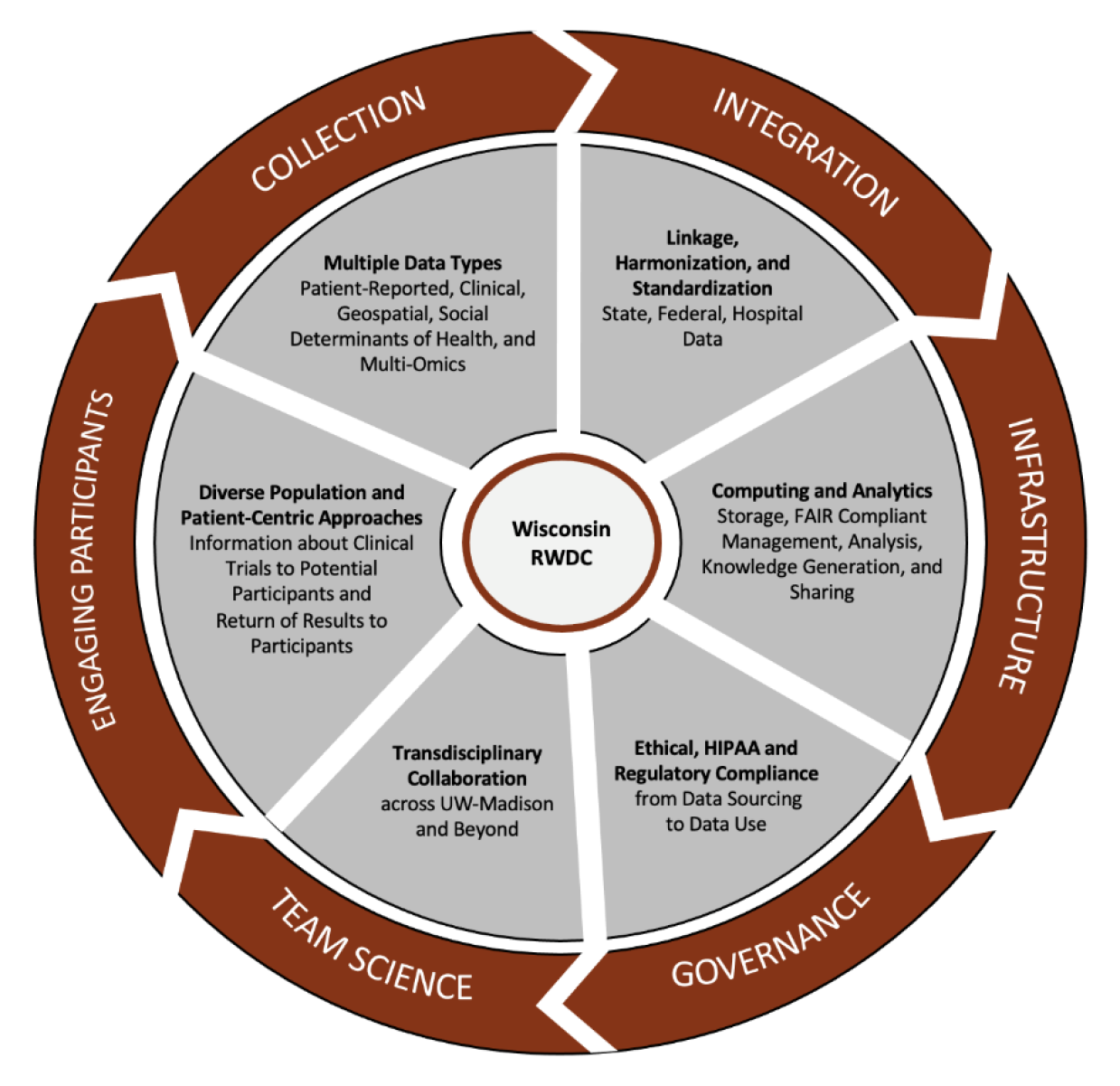
UW-Madison is developing a unique collaborative to enable ethical sourcing, standardization, quality control, annotation, integration, and analysis of biomedical data in a privacy-compliant environment. The Wisconsin Real-World Data Collaborative (RWDC) addresses several major challenges with real-world data:
- Incompleteness of data: A single EHR often does not include complete health care information generated over a patient’s life course, as patients move among places, medical facilities, and pharmacies. In addition, both genetic and exposure factors may be missing or difficult to access in EHRs. Data linkage across several complementary data sources is a common method to compensate for this incompleteness of EHR data. Given the utmost importance of patient privacy, research ethics, and compliance, we are implementing EHR data linkage with state and national data resources using privacy-preserving linkage tools.7 Our approaches promise to safely access data from multiple sources.
- Small sample size: When an academic medical center does not have enough patients to answer a research question—as in the case of a rare disease—collaborative integration of data from multiple sites becomes necessary. Such integration is often challenging due to local differences in data definitions. The Wisconsin RWDC is designed to map and harmonize data to common data models and vocabularies.
- Lack of diversity in research and inadvertent bias: Many academic medical centers have limited diversity in their patient populations. This can result in bias in sample selection and a lack of generalizability of findings across communities, states, and the country. The SMPH’s Survey of the Health of Wisconsin (SHOW)—funded by the Wisconsin Partnership Program (WPP)—effectively addresses these issues through its statewide, randomly selected cohort for biomedical research. Between 2008 and 2019, the program enrolled more than 6000 state residents (including children) with well-characterized health and health outcomes data. SHOW’s biorepository includes approximately 210,000 samples of urine, stool, blood, and derivative samples collected from participants who consented to use of their data in future research projects. SHOW uses advanced survey methods to ensure inclusion of participants that represent the diverse populations of Wisconsin.8 In 2018-2019, intentional oversampling was applied to include disadvantaged and hard-to-reach populations that otherwise would not be represented in biomedical research; one-third of the sample includes residents of rural areas.
With a follow-up survey response rate of more than 60%, the cohort presents a remarkable opportunity to engage a diverse community of participants in prevention and treatment studies. SHOW is being incorporated into the Wisconsin RWDC to accelerate availability of harmonized data and annotated specimens for research.
With heterogenous, real-world data and a Wisconsin-centric cohort, the Wisconsin RWDC (Figure) will evolve into a unique data repository for innovative research.
Innovation with Platform X and Data Science Dry Lab Suites
Data-driven biomedical research relies heavily on access to computing environments in which data can be securely retrieved from multiple sources, integrated, and rapidly analyzed. Lack of scalability and cost-effectiveness of storage and computing often impair researchers’ abilities to analyze large data sets, particularly using analytical approaches such as machine learning and artificial intelligence. At the SMPH, our transformative Platform X computing environment features advanced data security, reliability, and scalability. Secure transfer of raw data, including protected health information, into Platform X and transfer of deidentified results out of the platform are facilitated through tools that enable authentication, authorization, and audit trails. To achieve reproducibility in data analytics, we also are developing the Data Science Dry Lab (DSDL) Suites, which bundle servers, reference datasets, analytical software, data pipelining tools, and algorithms commonly used to support each domain of biomedical research. The first of these innovative suites is supporting clinical data analysis and neuroimaging research for a large, multisite study.9
Future Vision
Emerging opportunities for expanding the capacity of research that utilizes large data sets need new systems and technologies for harnessing data in order to deliver their full potential benefits. Platform X, the Wisconsin RWDC, and the DSDL Suites are examples of new tools that can allow researchers to define phenotypes, obtain and analyze holistic longitudinal data, and download deidentified results. At the same time, we must engage all of the diverse populations within our state and communicate results to the participants. We are fully committed to the development of an inclusive, real-world data infrastructure that will accelerate advances in research, education, and clinical care, and in doing so improve the health of the residents of Wisconsin and beyond.
REFERENCES
- American Recovery and Reinvestment Act of 2009, Pub L 111-5, 123 Stat 115 (2009). https://www.govinfo.gov/content/pkg/PLAW-111publ5/pdf/PLAW-111publ5.pdf
- WHO Coronavirus (COVID-19) Dashboard. World Health Organization. Accessed Nov 11, 2022. https://covid19.who.int
- 1918 Pandemic (H1N1 virus). Centers for Disease Control and Prevention. Last reviewed March 20, 2019. Accessed Nov 18, 2022. https://www.cdc.gov/flu/pandemic-resources/1918-pandemic-h1n1.html
- Collins FS, Doudna JA, Lander ES, Rotimi CN. Human molecular genetics and genomics–important advances and exciting possibilities. N Engl J Med. 2021;384(1):1-4. doi:10.1056/NEJMp2030694 (2021).
- Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18(1):83. doi:10.1186/s13059-017-1215-1
- U.S. Food and Drug Administration. Considerations for the Use of Real-World Data and Real-World Evidence to Support Regulatory Decision-Making for Drug and Biological Products: Draft Guidance for Industry. December 2021. Accessed Nov 18, 2022. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/considerations-use-real-world-data-and-real-world-evidence-support-regulatory-decision-making-drug
- Kho AN, Cashy JP, Jackson KL, et al. Design and implementation of a privacy preserving electronic health record linkage tool in Chicago. J Am Med Inform Assoc. 2015;22(5):1072-1080. doi:10.1093/jamia/ocv038
- Malecki KMC, Nikodemova M, Schultz A, et al. The Survey of the Health of Wisconsin (SHOW) Program: An infrastructure for advancing population health. Front Public Health. 2022;10:818777. doi:10.3389/fpubh.2022.818777
- National Institutes of Health. Neighborhood Socioeconomic Contextual Disadvantage and Alzheimer’s Disease. Accessed Nov 11, 2022. https://grantome.com/grant/NIH/RF1-AG057784-01